Step3_Cell_scoring
Last updated: 2021-08-20
Checks: 7 0
Knit directory: SCENIC_pipeline/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210818) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0110fe0. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/_site.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Step3_Cell_scoring.Rmd) and HTML (docs/Step3_Cell_scoring.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 728c832 | lily123920 | 2021-08-18 | diyici |
| html | 728c832 | lily123920 | 2021-08-18 | diyici |
该分析环节简介
背景:以上分析已经获得了该细胞群所涉及的可信的regulons(TF-targets)。但是regulons可能涉及数百个(length(table(regulonTargetsInfo$TF))#319该分析涉及319个)。。。并不是所有的regulons都是我们感兴趣的,那么如何挑选出感兴趣的或者说与研究表型相关的regulons呢?
策略:以TF为出发点,即以基因集为单位,评估每个细胞的每个regulon的活性,获得regulon活性矩阵。
实现方法:基于AUCell包的算法, 一种识别具有活跃基因调控网络细胞的新方法。
①输入:gene set或regulon ②数据处理过程:基于recovery analysis的打分方法。以“每个细胞的每个regulon”为分析单位。 ③过程:对基因按照表达水平从高到底排序,统计每个表达值的基因数量,计算曲线下面积。 ④输出:一个AUCscore的矩阵,储存每个细胞的每个regulon activity(AUC)。 结果:得到两个矩阵: ①一个AUC socre矩阵(绝对打分); ②一个binary 矩阵(手动或自动设置阈值;标记on or off两个状态)
结果解读——AUC两层含义: 原则:评分的基础是基因的表达值,分数越高代表基因集的激活程度越高。 ①绝对意义:代表签名基因集(regulon内位于高表达一端的基因子集)的基因占比。 ②细胞内部比较:与同一个细胞内的其他regulon相比,该regulon的相对表达水平。 值越大,代表该regulon的targets 基因表达水平较高。 ③细胞之间:对于同一个regulon的不同细胞,该regulon的相对表达水平。AUC值越大,代表该regulon在该细胞的活性越高。
数据挖掘方向: ①对细胞进行聚类。。。(直接将AUC矩阵视为连续性变量作为输入)。 ②同一regulon在不同细胞的差异分析。。。(直接将AUC矩阵视为连续性变量作为输入) ③对于二元矩阵,可以热图可视化。。。
问题: 可不可以选择一个细胞内AUC值top10的regulon呢?即挑选基因表达水平较高的基因? 如何将regulon信息和基因的差异表达信息联合起来分析呢?
一、代码
说明:为了节约计算资源,推断regulon的时候随机筛选了1000个细胞。。。但是对细胞进行regulon评分的时候,可以针对所有的细胞。 原理:前面是推断regulon,本质是从细胞群中识别出重要的TF-Targets关系【1000个细胞推断的结果可能可靠,也可能不可靠哦】。 而评分的基础是基因表达值。
1. regulon活性打分
##==regulon活性评分与可视化==##
##regulons计算AUC值并进行下游分析
scenicOptions1 <- initializeScenic(org="mgi",
nCores=1, # 这里不支持多线程,需要临时设置nCores = 1
dbDir=mydbDIR,
dbs = mydbs,
datasetTitle = "first try")
exprMat_all <- exprMat
exprMat_all <- log2(exprMat_all+1)
runSCENIC_3_scoreCells(scenicOptions1, exprMat=exprMat_all) 2. 构建regulon活性的binary矩阵
因为涉及到阈值调整,所以可以在通过regulon差异表达分析等策略筛选出想去的regulon后进行二进制的转化和探索及可视化。 通过互动的方式设置AUC阈值。。。。
# 探索结果
# 3.4 为最重要结果:行为regulon,列为细胞。
regulonAUC <- readRDS("./int/3.4_regulonAUC.Rds")
data <- regulonAUC@assays@data@listData[["AUC"]]
threshold <- readRDS("./int/3.5_AUCellThresholds.Rds")
#使用shiny互动调整阈值
aucellApp <- plotTsne_AUCellApp(scenicOptions, exprMat_all)
savedSelections <- shiny::runApp(aucellApp)
#保存调整后的阈值
newThresholds <- savedSelections$thresholds
scenicOptions@fileNames$int["aucell_thresholds",1] <- "int/newThresholds.Rds"
saveRDS(newThresholds, file=getIntName(scenicOptions, "aucell_thresholds"))
saveRDS(scenicOptions, file="int/scenicOptions.Rds")
# 二进制转换及衍生分析
runSCENIC_4_aucell_binarize(scenicOptions, exprMat=exprMat_all)二、结果文件
runSCENIC_3_scoreCells()是一个多种功能打包的函数,它包含的子功能有:
计算regulon在每个细胞中AUC值,最后得到一个以regulon为行细胞为列的矩阵。 结果目录:int/3.4_regulonAUC.Rds
计算每个regulon的AUC阈值,细胞中regulonAUC值>阈值,代表此regulon在细胞中处于激活状态,否则代表沉默状态。 结果目录:int/3.5_AUCellThresholds.Rds
使用regulonAUC矩阵对细胞进行降维聚类
用heatmap图展示regulonAUC矩阵,用t-SNE图分别展示每个regulon的活性分布情况。 结果目录:output/Step3_开头的系列文件
三、总结
SCENIC涉及的最主要的三个数据分析之旅co-expression module construction → 基于motif的 module 修剪 → 细胞评分到此就告一段落。。。最终有两个表非常重要,在后续数据挖掘的过程中会被反复使用,为:
①./output/Step2_RegulonTargetsInfo.tsv
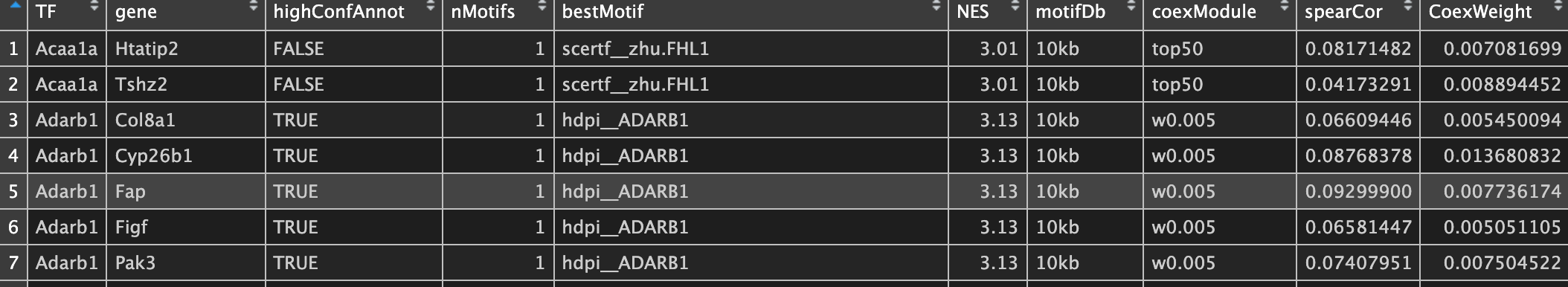
② regulon 活性矩阵 int/3.4_regulonAUC.Rds
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=Chinese (Simplified)_China.936
[2] LC_CTYPE=Chinese (Simplified)_China.936
[3] LC_MONETARY=Chinese (Simplified)_China.936
[4] LC_NUMERIC=C
[5] LC_TIME=Chinese (Simplified)_China.936
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 whisker_0.4 knitr_1.33 magrittr_2.0.1
[5] R6_2.5.0 rlang_0.4.11 fansi_0.5.0 stringr_1.4.0
[9] tools_4.0.2 xfun_0.24 utf8_1.2.1 git2r_0.28.0
[13] jquerylib_0.1.4 htmltools_0.5.1.1 ellipsis_0.3.2 rprojroot_2.0.2
[17] yaml_2.2.1 digest_0.6.27 tibble_3.1.2 lifecycle_1.0.0
[21] crayon_1.4.1 later_1.2.0 sass_0.4.0 vctrs_0.3.8
[25] promises_1.2.0.1 fs_1.5.0 glue_1.4.2 evaluate_0.14
[29] rmarkdown_2.9 stringi_1.5.3 bslib_0.2.5.1 compiler_4.0.2
[33] pillar_1.6.1 jsonlite_1.7.2 httpuv_1.6.1 pkgconfig_2.0.3