Step0_Data_preparation
Last updated: 2021-08-20
Checks: 7 0
Knit directory: SCENIC_pipeline/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210818) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0110fe0. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/_site.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Step0_Data_preparation.Rmd) and HTML (docs/Step0_Data_preparation.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 728c832 | lily123920 | 2021-08-18 | diyici |
| html | 728c832 | lily123920 | 2021-08-18 | diyici |
设置工作路径
创建新文件夹SCENIC,并将工作目录设置到./SCENIC/下。
准备工作目录

一、准备meta信息
meta信息主要是为了后续的数据挖掘与可视化,在共表达网络构建及细胞regulon评分过程不涉及该数据。
## 准备meta信息
load("~/sce_all_celltype.RData")
DefaultAssay(sce) <- "RNA"
sce_used <- sce[,sce$celltype %in% c("Modulated SMC", "VSMC") & sce$genotype == "wt"]
cellInfo <- sce_used@meta.data
colnames(cellInfo)[which(colnames(cellInfo)=="orig.ident")] <- "sample"
colnames(cellInfo)[which(colnames(cellInfo)=="SCT_snn_res.0.5")] <- "cluster"
colnames(cellInfo)[which(colnames(cellInfo)=="celltype")] <- "celltype"
cellInfo <- cellInfo[,c("sample","cluster","celltype")]
save(cellInfo, file="int/cellInfo.RData")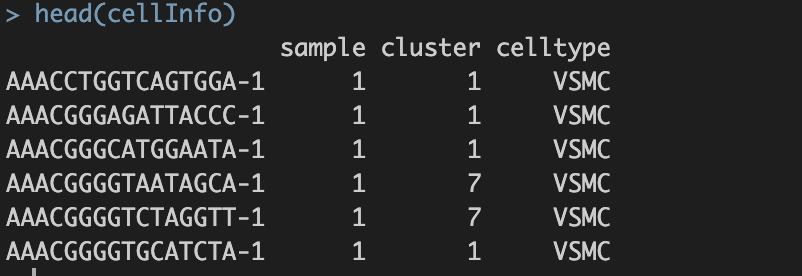
CellInfo数据格式示例:行名——细胞名;列名——需要的meta信息
二、准备表达矩阵
后续在共表达网络构建及细胞评分环节,有两个步骤非常占用计算资源。
测试流程时选用1000个细胞,runGenie3(exprMat_filtered_log, scenicOptions, nParts = 20) 耗时4个小时;runSCENIC_2_createRegulons(scenicOptions) 耗时2.5小时。
表达矩阵数据类型: 1. 优先选择count矩阵,read count和UMI count均可。 2. TPM或FPKM或RPKM也可。尽管有的作者会担心使用校正后的数据会引入artificial 共表达关系,但是在SCENIC团队的测试中对结果无影响。【2007;natrue methods.】
## 准备表达矩阵
#为了节省计算资源,随机抽取1000个细胞的数据子集。 # 实际数据分析中无需这个步骤
# subcell <- sample(colnames(sce_used),1000)
# sce_used <- sce_used[,subcell]
#save(sce_used, file = "sce_for_SCENIC.RData")
exprMat <- as.matrix(sce_used@assays$RNA@counts)
saveRDS(exprMat, file="int/exprMat.RData")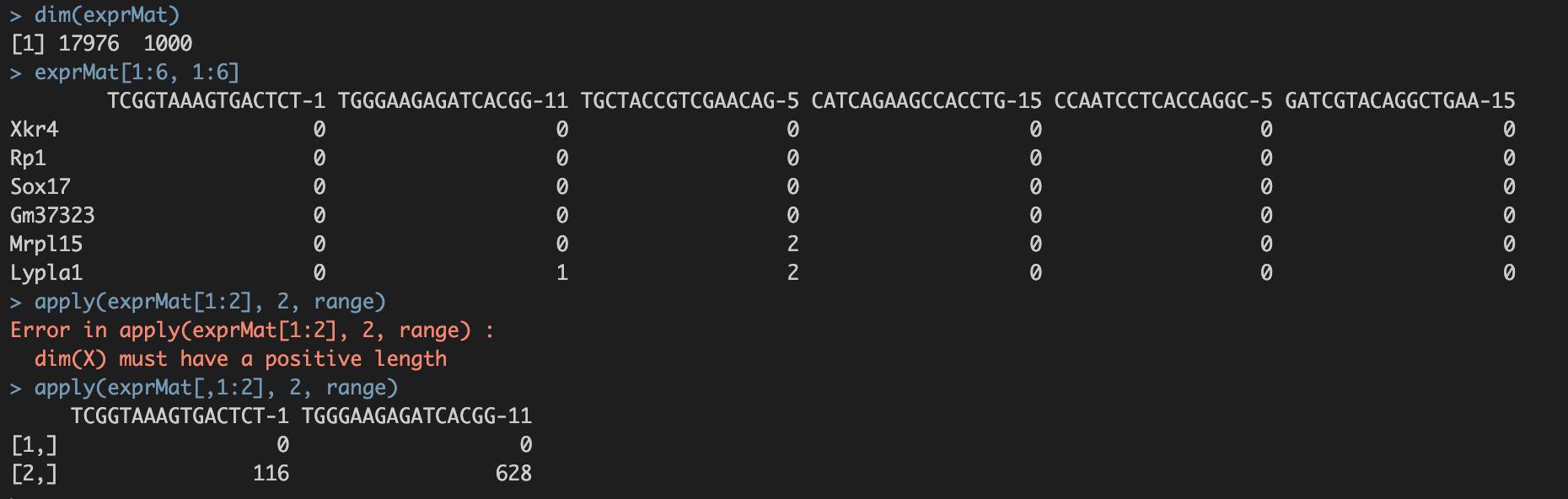
exprMat数据格式示例:行名——基因名;列名——细胞名;value——优推counts值
三、准备数据库
1. 数据下载路径
1.1, For human:
dbFiles <- c("https://resources.aertslab.org/cistarget/databases/homo_sapiens/hg19/refseq_r45/mc9nr/gene_based/hg19-500bp-upstream-7species.mc9nr.feather",
"https://resources.aertslab.org/cistarget/databases/homo_sapiens/hg19/refseq_r45/mc9nr/gene_based/hg19-tss-centered-10kb-7species.mc9nr.feather")
# mc9nr: Motif collection version 9: 24k motifs
1.2, For mouse:
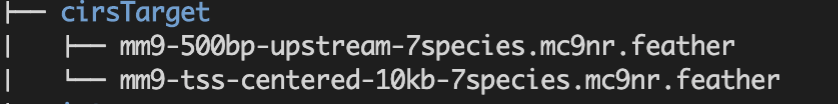
dbFiles <- c("https://resources.aertslab.org/cistarget/databases/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-500bp-upstream-7species.mc9nr.feather",
"https://resources.aertslab.org/cistarget/databases/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-tss-centered-10kb-7species.mc9nr.feather")
# mc9nr: Motif collection version 9: 24k motifs
1.3, For fly:
dbFiles <- c("https://resources.aertslab.org/cistarget/databases/drosophila_melanogaster/dm6/flybase_r6.02/mc8nr/gene_based/dm6-5kb-upstream-full-tx-11species.mc8nr.feather")
# mc8nr: Motif collection version 8: 20k motifs2. 下载方式
2.1 终端下载
将路径设置"SCENIC/cirsTarget"到在终端运行wget -c 路径名,下载相应的数据库文件。
2.2 Rstudio内下载
{r eval=FALSE, include=TRUE}
# 将工作目录设置”/cirsTarget“。。。不要忘了将路径返回到“SCENIC”下哦。
#如果3个参考数据库都想下载,每次设置变量dbFiles后,都要运行以下代码
for(featherURL in dbFiles)
{
download.file(featherURL, destfile=basename(featherURL)) # saved in current dir
}
四、设置分析环境
在数据分析之前,首先要配置好分析环境。主要涉及以下几个方面:
- org:注明使用的物种。有三个选择:
- nCores:使用的线程数,根据电脑CPU情况而定;
- dbDir:数据库存放目录,需要构建新的文件夹。此处为“
./SCENIC/cirsTarget”; - dbs:数据文件的名称
- datasetTitle:随意就好。
因此,有三个参数的设置要非常重视:org, dbDir, dbs。【一定不能错哦!】
这三个参数相当于告诉计算机,从motif信息数据储存在cirsTarget文件夹下,名称为……。这样计算机就知道去哪里调用数据了。
##设置分析环境
mydbDIR <- "./cirsTarget"
mydbs <- c("mm9-500bp-upstream-7species.mc9nr.feather",
"mm9-tss-centered-10kb-7species.mc9nr.feather")
names(mydbs) <- c("500bp", "10kb")
scenicOptions <- initializeScenic(org="mgi",
nCores=8,
dbDir=mydbDIR,
dbs = mydbs,
datasetTitle = "first try")
save(scenicOptions, "int/scenicOptions.RData")总结
这样,我们就准备好了后续分析使用的三个数据,并确保①exprMat和cellInfo数据格式正确;②database储存在正确路径下;③分析环境配置正确,并存储在scenicOption中,方便后续调用。
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=Chinese (Simplified)_China.936
[2] LC_CTYPE=Chinese (Simplified)_China.936
[3] LC_MONETARY=Chinese (Simplified)_China.936
[4] LC_NUMERIC=C
[5] LC_TIME=Chinese (Simplified)_China.936
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 whisker_0.4 knitr_1.33 magrittr_2.0.1
[5] R6_2.5.0 rlang_0.4.11 fansi_0.5.0 stringr_1.4.0
[9] tools_4.0.2 xfun_0.24 utf8_1.2.1 git2r_0.28.0
[13] jquerylib_0.1.4 htmltools_0.5.1.1 ellipsis_0.3.2 rprojroot_2.0.2
[17] yaml_2.2.1 digest_0.6.27 tibble_3.1.2 lifecycle_1.0.0
[21] crayon_1.4.1 later_1.2.0 sass_0.4.0 vctrs_0.3.8
[25] promises_1.2.0.1 fs_1.5.0 glue_1.4.2 evaluate_0.14
[29] rmarkdown_2.9 stringi_1.5.3 bslib_0.2.5.1 compiler_4.0.2
[33] pillar_1.6.1 jsonlite_1.7.2 httpuv_1.6.1 pkgconfig_2.0.3