Overview_of_SCENIC_workflow
Last updated: 2021-08-20
Checks: 2 0
Knit directory: SCENIC_pipeline/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0110fe0. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/_site.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Overview_of_SCENIC_workflow.Rmd) and HTML (docs/Overview_of_SCENIC_workflow.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 728c832 | lily123920 | 2021-08-18 | diyici |
| html | 728c832 | lily123920 | 2021-08-18 | diyici |
一、SCENIC简介
SCENIC,Single-cell rEgulatory Network Inference and Clustering,专为单细胞数据开发的GRNs算法,旨在识别高可靠性的由转录因子主导的的GRNs。
1. 提出的生物学背景
组织内细胞异质性的基础是细胞转录状态的差异,转录状态的特异性又是由转录因子主导的基因调控网络GRNs决定并维持稳定的。
2. 提出的组学背景
单细胞转录组数据具有背景噪音高、基因检出率低和表达矩阵稀疏性的特点,给传统的统计学和生物信息学方法推断高质量的GRNs带来了挑战。
3. SCENIC优势
专为单细胞数据开发的GRNs算法,创新之处在于引入了转录因子motif序列验证统计学方法推断的基因共表达网络。旨在识别高可靠性的由转录因子主导的的GRNs。
二、SCENIC工作流程概览
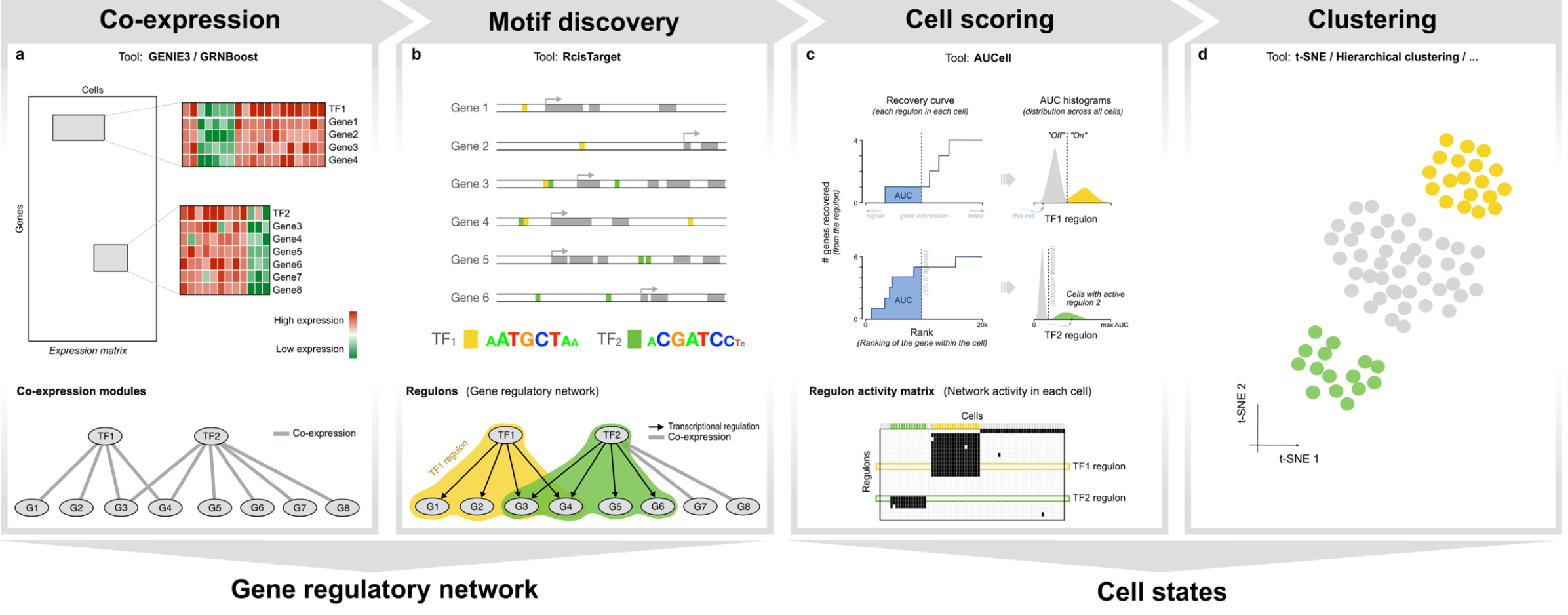
| 步骤 | 包基础 | 输入数据 | 算法核心 | 输出数据 | 完成任务 | 涉及概念 | |
|---|---|---|---|---|---|---|---|
| 核心步骤——数据分析处理 | Step1 | GENIE3/GRNBoost | 表达矩阵 | 以TF为出发点,基于共表达情况鉴定每个TF的潜在靶点 |
module/network/ | ||
| 核心步骤——数据分析处理 | Step2 | RcisTarget | 基于DNA-motif分析选择潜在的直接结合靶点 |
regulon | |||
| 核心步骤——数据分析处理 | Step3 | AUCell | AUC | 通过打分评估细胞内不同转录因子的调控活性;或者说分析每个细胞的regulons活性 |
|||
| 数据探索和可视化 | Step4 | t-SNE/Hierarchical clustering/…… | 基于regulons的活性鉴定稳定的细胞状态并对结果进行探索 |
三、参考资料
想要深入了解分析原理和流程,可以参考以下两篇文献:
SCENIC : single-cell regulatory network inference and clustering (2007年首先发表于nature methods)
A scalable SCENIC workflow for single-cell gene regulatory network analysis (2020年将重新整理后的流程发表于nature protocles)